
Yesterday I attended a talk on deep learning and genomics by Pi-Chauan Chang, a software engineer at Google. Pi-Chauan gave a high level overview of deep learning and how her team formulates a problem in genomics to successfully apply deep learning techniques. She also discussed DeepVariant – a software built by Google to enable community efforts to progress genomic sequencing.
What is deep learning?:
Deep Learning is a subfield of machine learning concerned with algorithms inspired by the structure and function of the brain called artificial neural networks.
Deep learning is playing a huge role in advancements in genomic research such as high processing of sequencing techniques. This information era where we continue to be presented with an outpouring of data has truly began to challenge conventional methods used in genomics. While deep learning has succeeded in a variety of fields such as vision, speech, and text processing it is now presented with the unique challenge of helping us to explore beyond our current knowledge to interpret the genome.
Pi-Chauan Chang shared that genome sequencing is a core technology in biology.. It allows us to ask how can we personalize medicine based on genome?
What is a Genome?:
A genome is an organism’s complete set of genetic instructions. Each genome contains all of the information needed to build that organism and allow it to grow and develop.
There are 23 chromosome we inherent from our parents.
Most of our DNA is similar.. 99.9% of our DNA are the same– this makes us human..
Its the .1 pecent that makes us unique.
The Human Genome Project was a milestone of genome sequencing . This was the massive international collaboration to map the complete human genome. This project outputted a genome dictionary ~ 3.2 million characters.
A decade ago it was expensive to sequence people..now it cost ~$1000 to sequence an individual. This creates much opportunity for precision medicine.
There is, however, a trade off.
The new sequencing technology has errors! From blood draw computational biologists get raw data(characters of ACTG) which are really short snippets of the whole genome.. much like puzzle pieces. They try to map the puzzle pieces but are faced having to find the variants.
Variant calling:
Variant calling is the process by which we identify variants from sequence data.
Typically variant calling consist of a three step process:
- Carry out whole genome or whole exome sequencing to create FASTQ files.
- Align the sequences to a reference genome, creating BAM or CRAM files.
- Identify where the aligned reads differ from the reference genome and write to a VCF file.

The audience was informed that it is pretty common that computational biologists regularly inspect genomic data..
The question at hand is can we teach machines to perform the same task? Can we teach a machine to detect the variants?
This is where deep learning steps in.
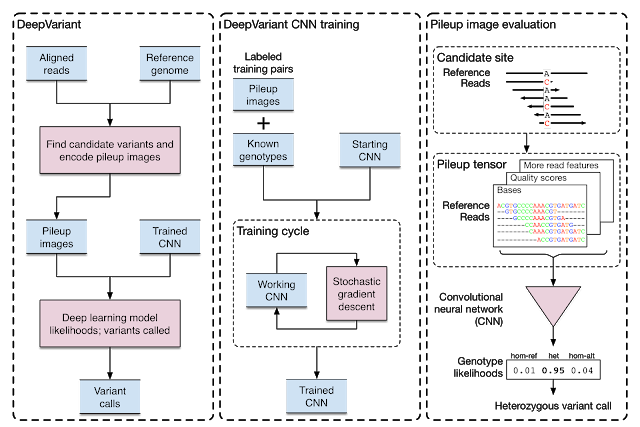
DeepVariant
DeepVariant is a deep learning technology to reconstruct the true genome sequence from HTS(high-throughput sequencing) sequencer data with significantly greater accuracy than previous classical methods. DeepVariant transforms the task of variant calling, as this reconstruction problem is known in genomics, into an image classification problem well-suited to Google’s existing technology and expertise.
DeepVariant is now an open source software to encourage collaboration and to accelerate the use of this technology to solve real world problems!
https://github.com/google/deepvariant